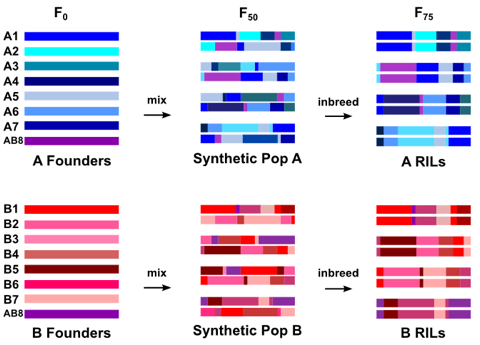
Creating the DSPR
The Recombinant Inbred Lines
The recombinant inbred lines (RILs) are derived from two synthetic populations. Each synthetic populations is derived from a set of 8 inbred
P-free lines representing a worldwide sample. For detailed information about the founder lines used to create the two synthetic populations (A and B), see the table below. The two synthetic populations share one line (AB8: Samarkand
ry506; obtained from Trudy Mackay, NC State) in common, thus the two populations are constructed from 15 lines overall. The two synthetic populations were created in Fall 2006. In the first generation, the 8 founder lines were crossed in a round-robin mating scheme to bring all founder alleles together. The populations were subsequently maintained as large, random-mating cohorts for a total of 50 generations, at which point the RILs were created. A set of 750 RILs was created from each synthetic population by 25 generations of full-sib mating. This design creates a panel of RILs in which the genome of each RIL is a fine-scale mosaic of segments from the 8 founder lines.
The Founder Lines
| Founder |
Vial Code |
Stock Center |
Stock Number |
Stock Name |
Details |
|
|
| A1 |
1 |
Bloomington |
1 |
Canton-S |
i18+ |
|
|
| A2 |
3841 |
Bloomington |
3841 |
BOG1 |
i18+ |
|
|
| A3 |
3844 |
Bloomington |
3844 |
BS1 |
i18+ |
|
|
| A4 |
3852 |
Bloomington |
3852 |
KSA2 |
i18+ |
|
|
| A5 |
3875 |
Bloomington |
3875 |
VAG1 |
i18+ |
|
|
| A6 |
3886 |
Bloomington |
3886 |
wild5B |
i18+ |
|
|
| A7 |
T.7 |
Tucson |
14021-0231.7 |
n/a |
i18+ |
|
|
| AB8 |
Sam |
TFC Mackay |
n/a |
Sam; ry506 |
+ |
|
|
| B1 |
3839 |
Bloomington |
3839 |
BER1 |
i18+, tetracyline treated |
|
|
| B2 |
3846 |
Bloomington |
3846 |
CA1 |
i18+ |
|
|
| B3 |
3864 |
Bloomington |
3864 |
QI2 |
i9+, tetracyline treated |
|
|
| B4 |
3870 |
Bloomington |
3870 |
RVC3 |
i3+ |
|
|
| B5 |
T.0 |
Tucson |
14021-0231.0 |
n/a |
i18+ |
|
|
| B6 |
T.1 |
Tucson |
14021-0231.1 |
n/a |
i18+ |
|
|
| B7 |
T.4 |
Tucson |
14021-0231.4 |
n/a |
i18+ |
|
|
|
| A, B |
Refers to which synthetic population(s) the line
founded. |
|
|
|
| i3, i9, i18 |
Refers to the # of generations of inbreeding on
arrival from the stock center. |
|
|
| + |
Refers to the fact that line was
re-initiationed (post-inbreeding) from 4 males and 4 vigin females. Each of
these 8 founding flies was genotyped for strain-specific, diagnostic SNP to
check stock integrity. |
|
|
| tetracyline |
Stock harbored Wolbachia,
which was cleared with two generations on
tetracyline-containing food. |
|
|
Whole Genome Re-sequencing of Founder Lines
Using Illumina paired-end 54bp sequencing, we generated 50X genome sequence coverage for each founder, aligning the millions of short reads to the
D. melanogaster reference sequence to create an alignment of the 15 founders.
Uncovering the mosaic structure of the RILS
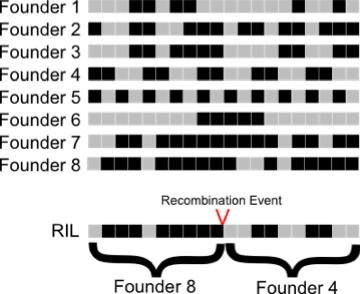
We used 96-plex RAD (restriction-site associated DNA markers) to discover and genotype ~10,000 SNP markers in the panel of RILs. This set of linked SNP markers allows us to uncover the mosaic founder structure of each RIL using a Hidden Markov Model (HMM) we have developed. The HMM uses this set of markers to determine the probability that each segment is derived from each of the 8 founder lines. While a single biallelic SNP can only distinguish between two allelic classes, a set of linked SNPs can distinguish between all 8 founders. This fact can be visualized in the example to the right where the pattern of markers (black and gray representing different alleles) allows identification of the founder ancestry. As SNP density increases, the level of genetic information also increases. The high SNP density used in the DSPR allows us to distinguish among the 8 founder lines with a high degree of confidence. The fact that we can infer the founder ancestry of each RIL chromosomal segment and have the full genome sequences of the founders, means we can essentially determine the full genome sequence of all the RILs with near perfect certainty.